“Freedom of design” in chemical compound space
Video recording:
 Speaker: Leonardo Medrano Sandonas (Department of Physics and Materials Science, University of Luxembourg)
Speaker: Leonardo Medrano Sandonas (Department of Physics and Materials Science, University of Luxembourg)
Title: “Freedom of design” in chemical compound space.
Time: Wednesday, 2023.06.14, 10:00 a.m. (CET)
Place: fully virtual (contact Jakub Lengiewicz to register)
Format: 30 min. presentation + 30 min. discussion
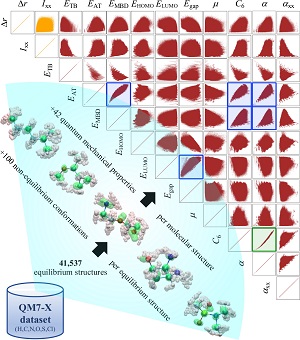
Abstract: The rational design of molecules with targeted quantum-mechanical (QM) properties requires an advanced understanding of the structure-property/property-property relationships that exist across chemical compound space (CCS). In particular, QM methods combined with machine learning (ML) techniques have accelerated the accurate calculation of molecular properties, providing a direct mapping from 3D molecular structure to property space. However, the development of reliable and efficient methodologies to define an inverse mapping is a long-standing challenge in organic chemistry that needs further investigation. Hence, in the present work, we address this challenge by probing the possibility of parameterizing the CCS with a finite set of extensive/intensive QM properties. In doing so, we design a proof-of-concept implementation based on a variational autoencoder (VAE) architecture and demonstrate that it is feasible to approximate such inverse mapping for small drug-like molecules contained in QM7-X dataset. By approaching the inverse design problem from this angle, we are not only able to retrieve the conditional generation of molecules with targeted properties, but we also open up a range of other possibilities which include the analysis of property-structure relationships as well as transition path interpolation. Our findings thus suggest a multifaceted route to understanding and manipulating organic molecules, and we hope that they also serve as motivation for the development of more interpretable ML models for inverse molecular design.
Leonardo Medrano is currently working as a postdoc in Prof. A. Tkatchenko’s group at the University of Luxembourg. He is combining machine learning methods with quantum/statistical mechanics to develop physics-inspired neural network potentials for the study of drug-protein interactions as well as frameworks for computer-aided molecular design. In 2018, Leonardo got his doctor degree in Mechanical Engineering at the Technical University of Dresden as an IMPRS fellow (International Max Planck Research School). Earlier, he got his bachelor and master’s degree in Physics at the National University of San Marcos in Lima-Peru. In addition to his theoretical investigations, Leonardo actively engages in multi-disciplinary projects with experimental/industrial collaborators to address current challenges in Physics and Chemistry (see Google Scholar page).